Al-powered modeling for generative ensembles
Conventional MD can help us sample protein conformations but it will be time- and resource-consuming to simulation adequately and accurately, especially for intrinsically distordered proteins (IDP). The conformational landscape of an IDP usually has many shallow “valleies” (states) and ideally one expects to sample all the conformational states and also the transitions between those states.
As you can image, IDPs are flexible and plastic and can adopt way more conformational states than those of folded proteins (where usually only a few significant states can account for function). This poses difficulties: we need to run simulations long enough to have adequate sampling that describe the conformational landscape properly and also enough number of transitions between different states (such that we can derive the hiddern free energy landscape). This is extremely challenging since not only it requires us to have an accurate and well-benchmarked IDP force field (believe me, this is very hard and people are still improving in this direction), but also it requires extremely long time simulation to populate all possible states and not to mention the transitions between different states (the relative probability densities of those states).
In this research direction, we propose to use deep learning neural networks to learn large MD simulation data of IDPs and hopeful can easily and quickly generate IDP ensembles with good qualities on the description of ensemble properties similar to MD ensembles. This can potentially avoid runing extremely long time simulations and predictively generate IDP ensembles for any given sequences.
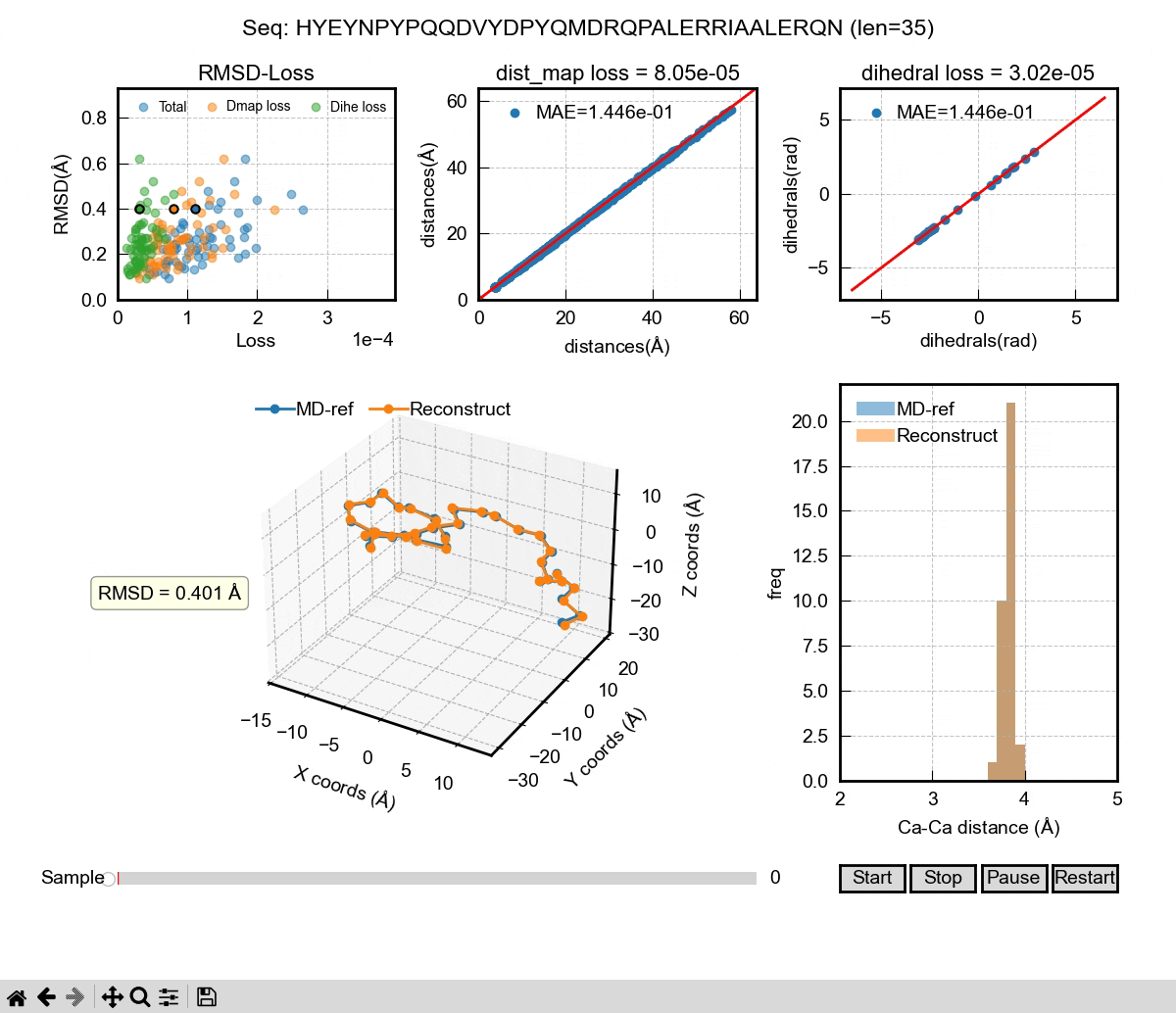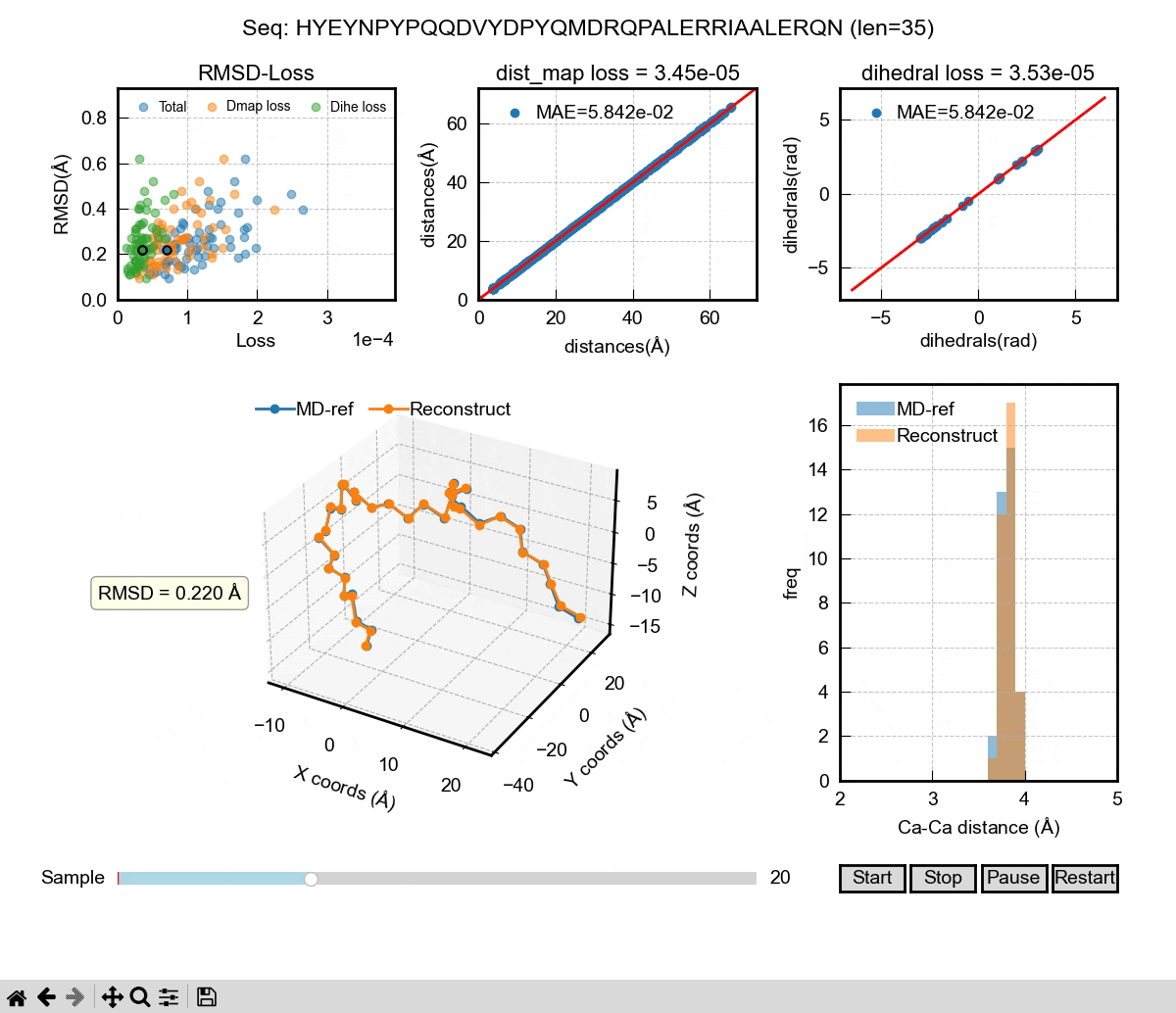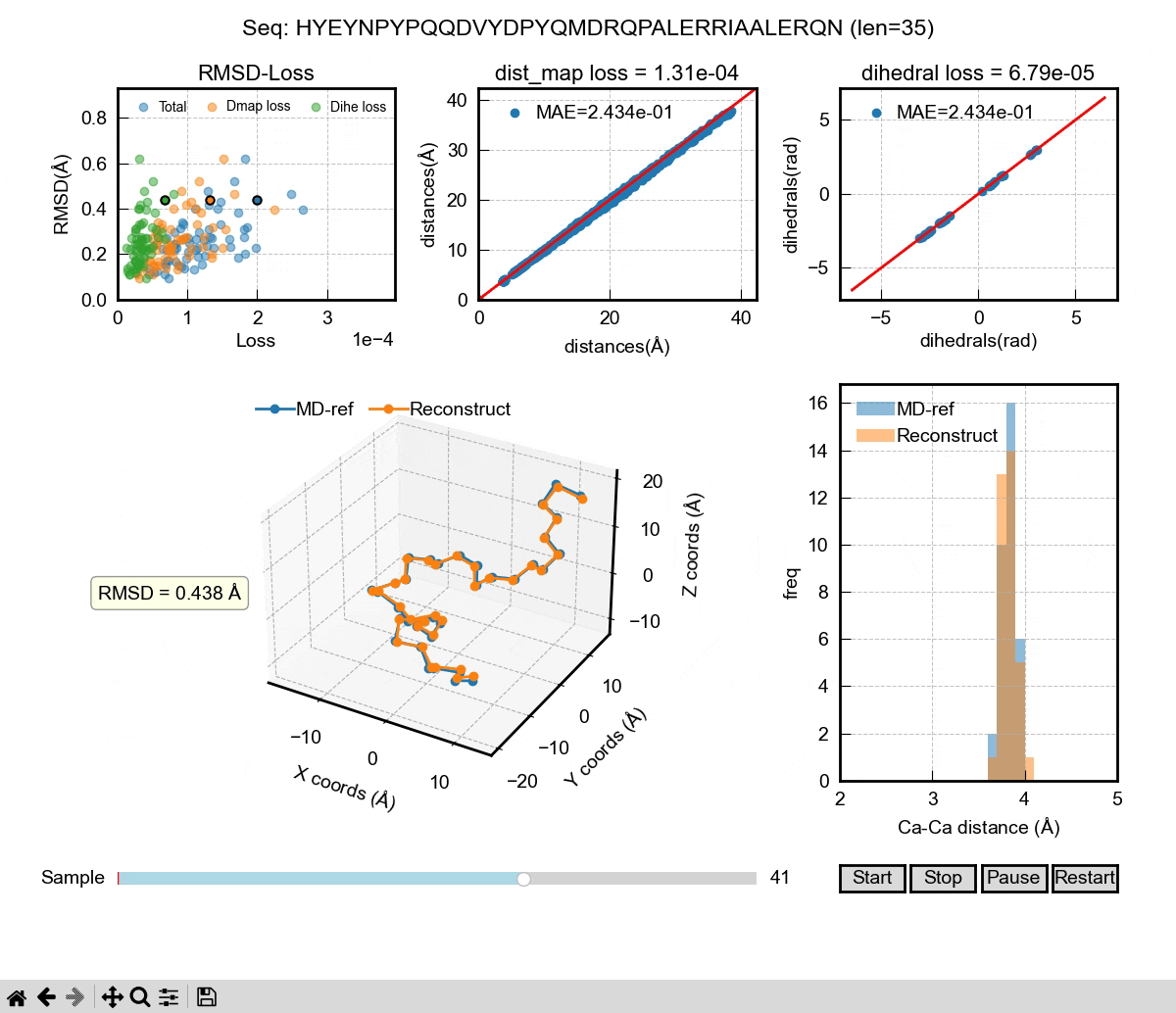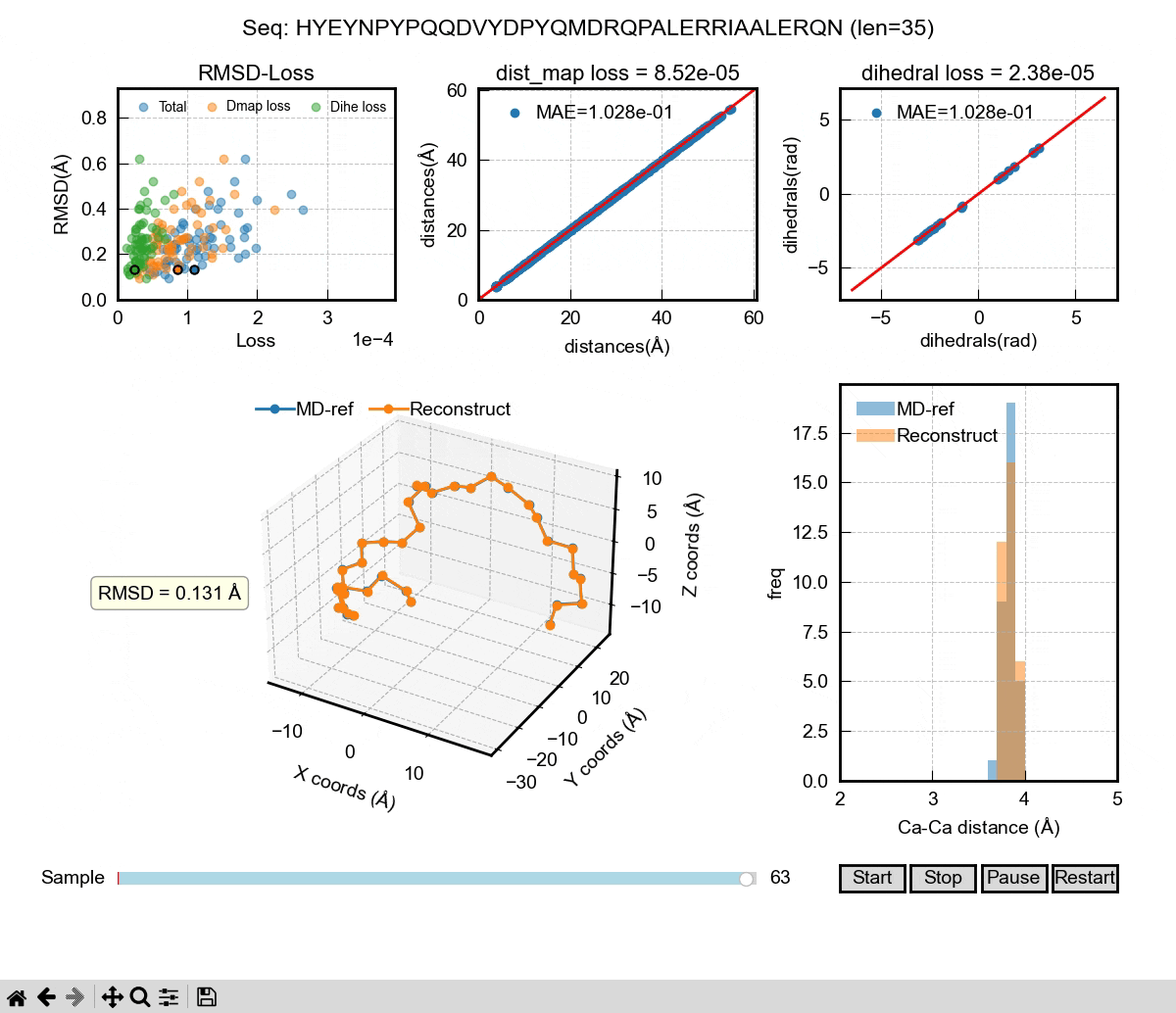
The above gif shows the first phase of our Al model – a autoencoder that can be used to reconstruct MD coordinates. As you can see in the 3D figure, each conformation can almost be perfectly reconstructed. We used dihedral angle loss and distance map loss in our training. The decoder of the autoencoder could be used to take a latent space representation and generate 3D coordinates. Then, it comes to the second phase of the Al model. We want to learn the latent space probability distributions of IDPs using the famous DDPM (denoising diffusion probabilistic model) architecture. It can learn from sequences and their latent space representations from the encoder in the training. In the inference, the DDPM will generate new latent space representations for a given sequence, which will then be decoded into 3D coordinates. If we generate enough frames of those coordinates, we can eventually have a Al generated IDP ensemble that hopefully captures the properties of a real MD ensemble, including Rg, secondary structure and other ensemble properties.
